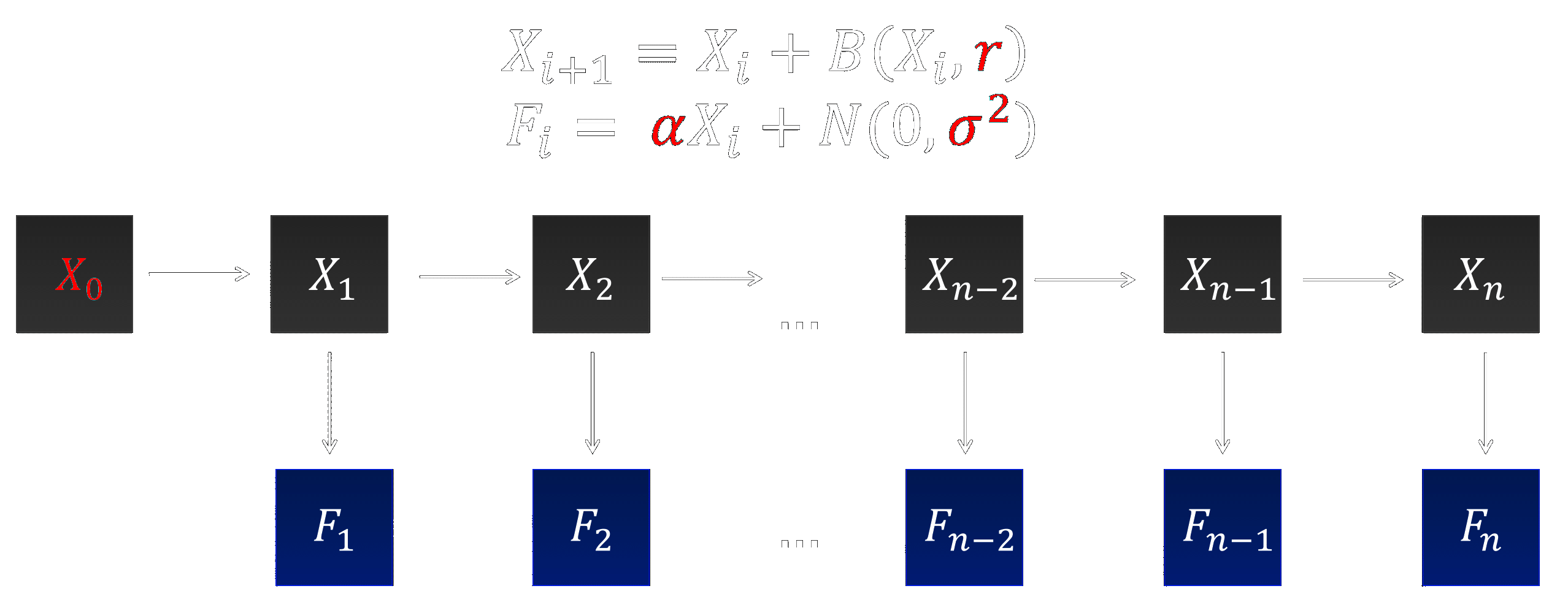Hidden Markov Model
Given fluorescence data from a qPCR experiment, the main target analyte we wish to infer is the initial mtDNA copy number $X_0$. Other model parameters may be inferred as a by-product of performing inference on $X_0$.
Lalam, 2007 proposes a binomial branching process combined with a HMM (Figure 8). It accounts for the fact that the number of replicated DNA molecules during qPCR is random, and additionally that the measurement of the fluorescence emitted by the DNA molecules is collected with some random error.For all $i\geq1$: $$X_{i+1} = X_i + B(X_i,r), \qquad F_i = \alpha X_i + N(0,\sigma^2)$$ where $X_i$ is the number of molecules and $F_i$ is the measured fluorescence at cycle $i$. Fluorescence reads are taken in the presence of background fluorescence, modelled as additive Gaussian noise with mean $0$ and variance $\sigma^2$. If the experimental data used for inference is not background corrected, a different mean and variance is required.
Parameters we wish to infer are:
- $X_0$: the initial number of molecules
- $r$: the efficiency of the qPCR experiment
- $\alpha$: the constant describing the proportionality of the fluorescence intensity emitted at a given cycle to the number of molecules at that point
- $\sigma$: the variance contributing to the noise term in the fluorescence expression